Epithelial plasma membrane
- Details
- Last Updated: Friday, 05 March 2021 13:23
Thanks to Stephen Fairweather, and the excellent work of O'Mara and coworkers, we are happy to share newly parameterized Martini models of lipids such as plasmalogens, hyrdoxylated sphingholipids, and Forssman lipids. For more information, check out the publication: https://doi.org/10.1063/5.0040887
You can download the topologies and other data from here!



























































































































































 An idealized brain plasma membrane composed of 58 different lipid types.
An idealized brain plasma membrane composed of 58 different lipid types.
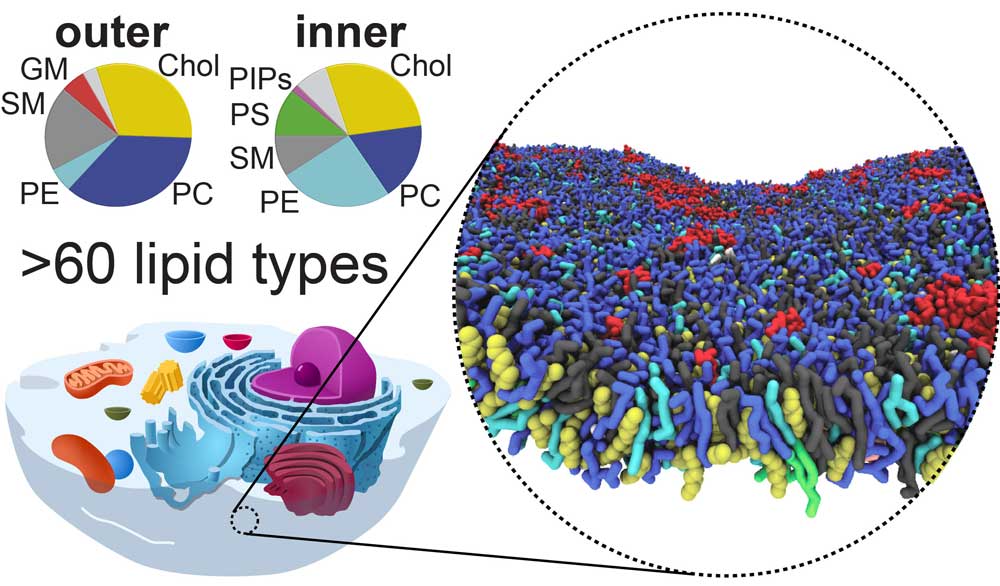 An idealized mammalian plasma membrane composed of 63 different lipid types.
An idealized mammalian plasma membrane composed of 63 different lipid types.