Simulating Protein Self-Assembly
- Details
- Last Updated: Wednesday, 25 January 2023 14:19
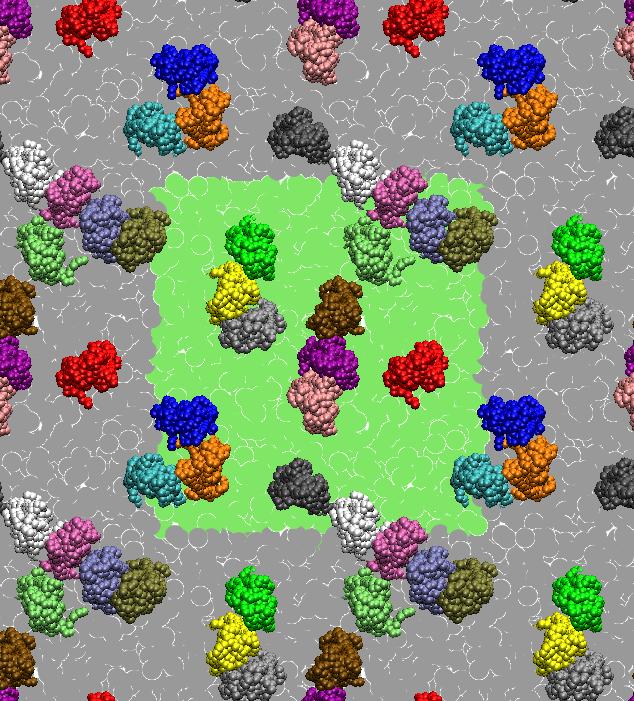
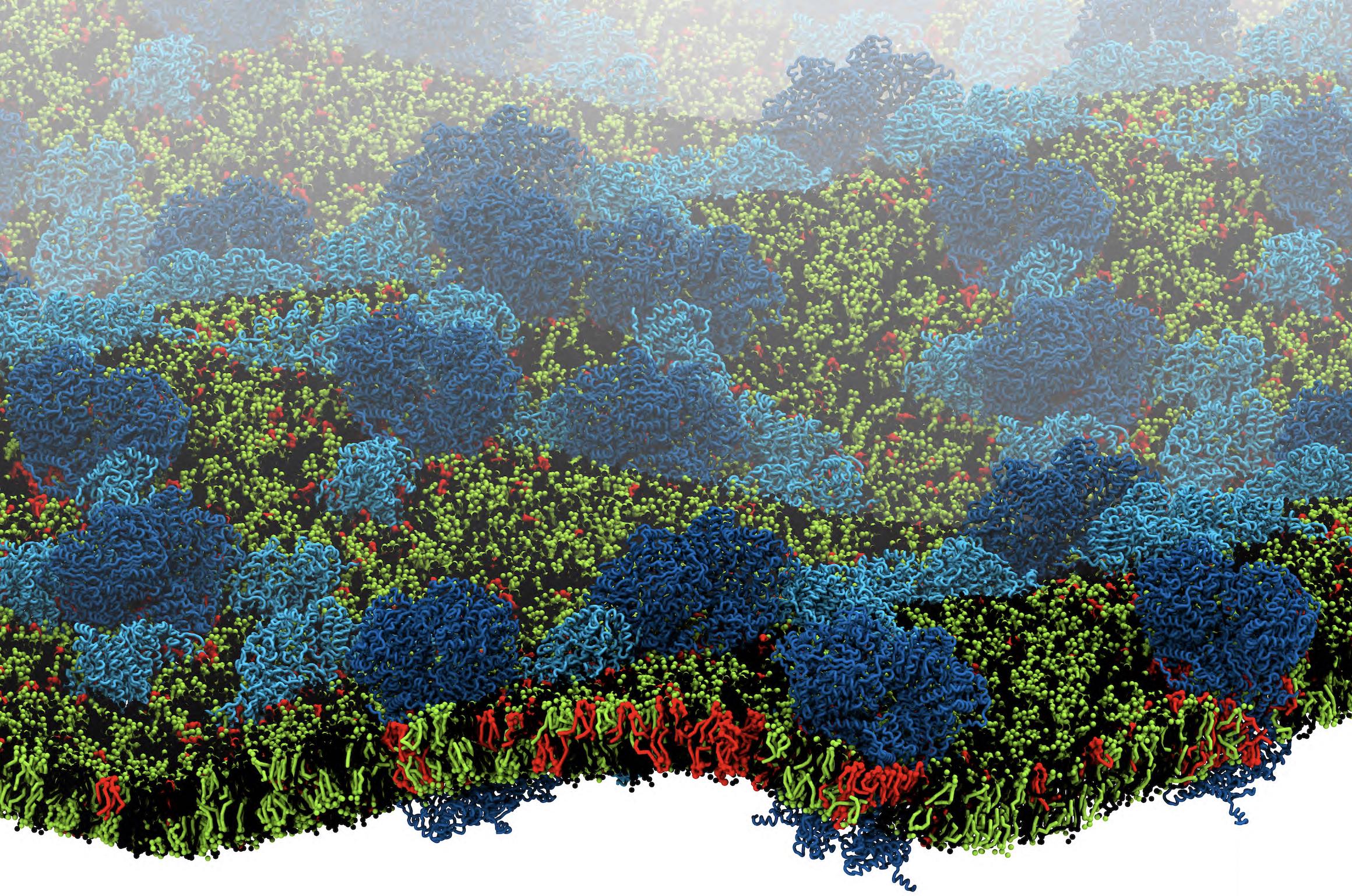
[1] X. Periole, T. Huber, S.J. Marrink, T. P. Sakmar. G protein-coupled receptors self-assemble in dynamics simulations of model bilayers. JACS, 129:10126-10132, 2007.
[2] D. Sengupta, S.J. Marrink. Lipid mediated Interactions tune the association of Glycophorin A helix and its disruptive mutants in membranes. Phys. Chem. Chem. Phys., 12:12987-12996, 2010. abstract
[3] X. Periole, M. Cavalli, S.J. Marrink, M. Ceruso. Combining an elastic network with a coarse-grained molecular force field: structure, dynamics and intermolecular recognition. J. Chem. Th. Comp., 5:2531-2543, 2009.
[4] D. Sengupta, A. Rampioni, S.J. Marrink. Simulations of the C-subunit of ATP-synthase reveal helix rearrangements. Mol. Membr. Biol., 26:422-434, 2009.
[5] L.V. Schafer, D.H. de Jong, A. Holt, A.J. Rzepiela, A.H. de Vries, B. Poolman, J.A. Killian, S.J. Marrink. Lipid packing drives the segregation of transmembrane helices into disordered lipid domains in model biomembranes. PNAS, 108:1343-1348, 2011. open access
[6] J. Domanski, S.J. Marrink, L.V. Schaefer. Transmembrane helices can induce domain formation in crowded model biomembranes. BBA Biomembr., in press, 2011. DOI:10.1016/j.bbamem.2011.08.021. abstract
[7] C.P. Chng, S.M. Tan. Leukocyte integrin αLβ2 transmembrane association dynamics revealed by coarse-grained molecular dynamics simulations. Proteins, 79:2203–2213, 2011.
[8] X. Periole, A.M. Knepp, T.P. Sakmar, S.J. Marrink, T. Huber. Structural determinants of the supra-molecular organization of G protein-coupled receptors in bilayers. JACS, 134:10959–10965, 2012. abstract
[9] J.M. Johnston, H. Wang, D. Provasi, M. Filizola. Assessing the relative stability of dimer interfaces in G protein-coupled receptors. PLoS Comput Biol 8: e1002649, 2012.
[10] Y. Komatsu, M. Fukuda, H. Yamada, S. Kawamoto, T. Miyakawa, R. Morikawa, M. Takasu, S. Yokojima, S. Akanuma, A. Yamagishi. Constructing protein nano-fiber and estimation of the electronic state around metal ions. Int. J. Quant. Chem. 112:3750–3755, 2012.
[11] I. Casuso, J. Khao, M. Chami, P. Paul-Gilloteaux, M. Husain, J.P. Duneau, H. Stahlberg, J.N. Sturgis, S. Scheuring. Characterization of the motion of membrane proteins using high-speed atomic force microscopy. Nat. Nanotechnology, in press, 2012. DOI: 10.1038/NNANO.2012.109.
[12] M. Baaden, S.J. Marrink. Coarse-grain modelling of protein–protein interactions. Curr. Opin. Struct. Biol., 23:878-886, 2013. open access
[13] H.I. Ingólfsson, C.A. Lopez, J.J. Uusitalo, D.H. de Jong, S. Gopal, X. Periole, S.J. Marrink. The power of coarse-graining in biomolecular simulations. WIREs Comput. Mol. Sci., 4:225–248, 2014. open access
[14] S.J. Marrink, V. Corradi, P.C.T. Souza, H.I. Ingolfsson, D.P. Tieleman, M.S.P. Sansom. Computational Modeling of Realistic Cell Membranes. Chem. Review, 119:6184–6226, 2019. doi:10.1021/acs.chemrev.8b00460
[15] T.A. Wassenaar, K. Pluhackova, A. Moussatova, D. Sengupta, S.J. Marrink, D.P. Tieleman, R.A. Böckmann. High-throughput simulations of dimer and trimer assembly of membrane proteins. The DAFT approach. JCTC, 11:2278–2291, 2015. abstract
[16] C. Arnarez, S.J. Marrink, X. Periole. Molecular mechanism of cardiolipin-mediated assembly of respiratory chain supercomplexes. Chem. Sci., 7:4435-4443, 2016. open access
[17] F. Sun, C.F.E. Schroer, L. Xu, H. Yin, S.J. Marrink, S.Z. Luo. Molecular Dynamics of the Association of L-Selectin and FERM Regulated by PIP2. Biophys. J., 114:1858–1868, 2018. doi:10.1016/j.bpj.2018.02.034
[18] F. Sun, C.F.E. Schroer, C.R. Palacios, L. Xu, S.Z. Luo, S.J. Marrink. Molecular mechanism for bidirectional regulation of CD44 for lipid raft affiliation by palmitoylations and PIP2. PLoS Comput. Biol. 16:e1007777, 2020. doi.org/10.1371/journal.pcbi.1007777
[19] I. Faustino, H. Abdizadeh, P.C.T. Souza, A. Jeucken, W.K. Stanek, A. Guskov, D.J. Slotboom, S.J. Marrink. Membrane mediated toppling mechanism of the folate energy coupling factor transporter. Nature Commun. 11:1763, 2020. doi.org/10.1038/s41467-020-15554-9
[20] J. Roel-Touris, C.G. Don, R.V. Honorato, J.P. Rodrigues, A.M.J.J. Bonvin. Less is more: Coarse-grained integrative modeling of large biomolecular assemblies with HADDOCK. J. Chem. Theory Comput. 15:6358-6367, 2019
[21] J. Roel-Touris, B. Jiménez-García, A.M.J.J. Bonvin. Integrative modeling of membrane-associated protein assemblies. Nature Communications 11:1-11, 2021.
[22] M.G. Chiariello, F. Grünewald, R. Zarmiento-Garcia, S.J. Marrink. pH-Dependent Conformational Switch Impacts Stability of the PsbS Dimer. J. Phys. Chem. Lett. 14, 905-911, 2023. doi:10.1021/acs.jpclett.2c03760


























































































































































