9: Polyply
- Details
- Last Updated: Sunday, 29 August 2021 09:08
In case of issues, please contact This email address is being protected from spambots. You need JavaScript enabled to view it.
This tutorial has three independent parts. It is recommended to start with part 1. However, people familiar with Martini can also go to part 2/3 independently. The tutorials cover applications and usage of Polyply, a python suite for facilitating simulations of (bio-)macromolecules and nanomaterials.1
Part 1: Melts and Blends
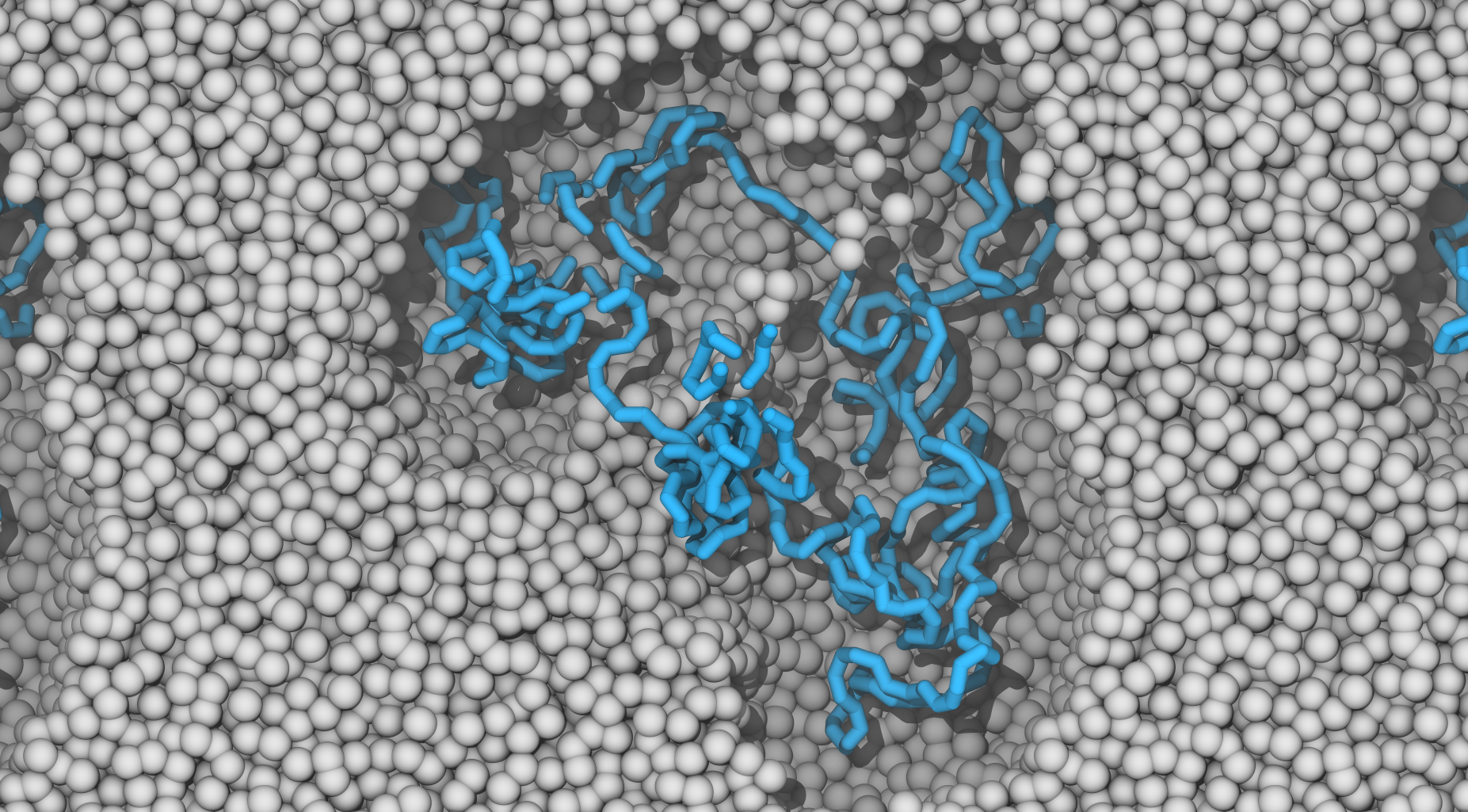
In this tutorial you will learn how to generate simple polymer melts and blends with polyply using Martini3. It takes about 10min to complete the Tutorial and covers the basic functionality of polyply itp file generation and coordinate generation. No prior knowledge is required to start this tutorial, but it is useful as background for the other two tutorials. Follow the link to get to the tutorial:
Part 2: PEGylated lipid bilayers
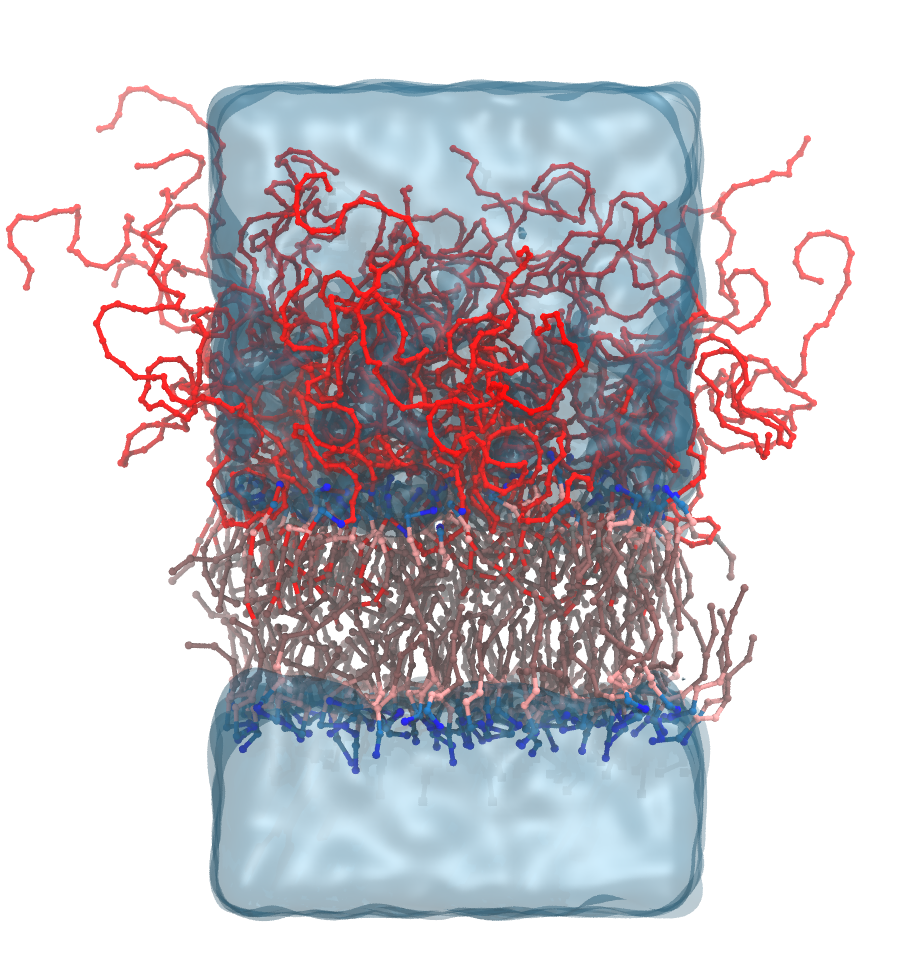
In this tutorial you will learn how to generate PEGylated lipid bilayers. It serves as an example tp explains how to connect a biomolecule to a polymer, build brushes, use pre-build coordinates from other programs. Follow the link to get to the tutorial:
Part 3: Early Life in a vesicle
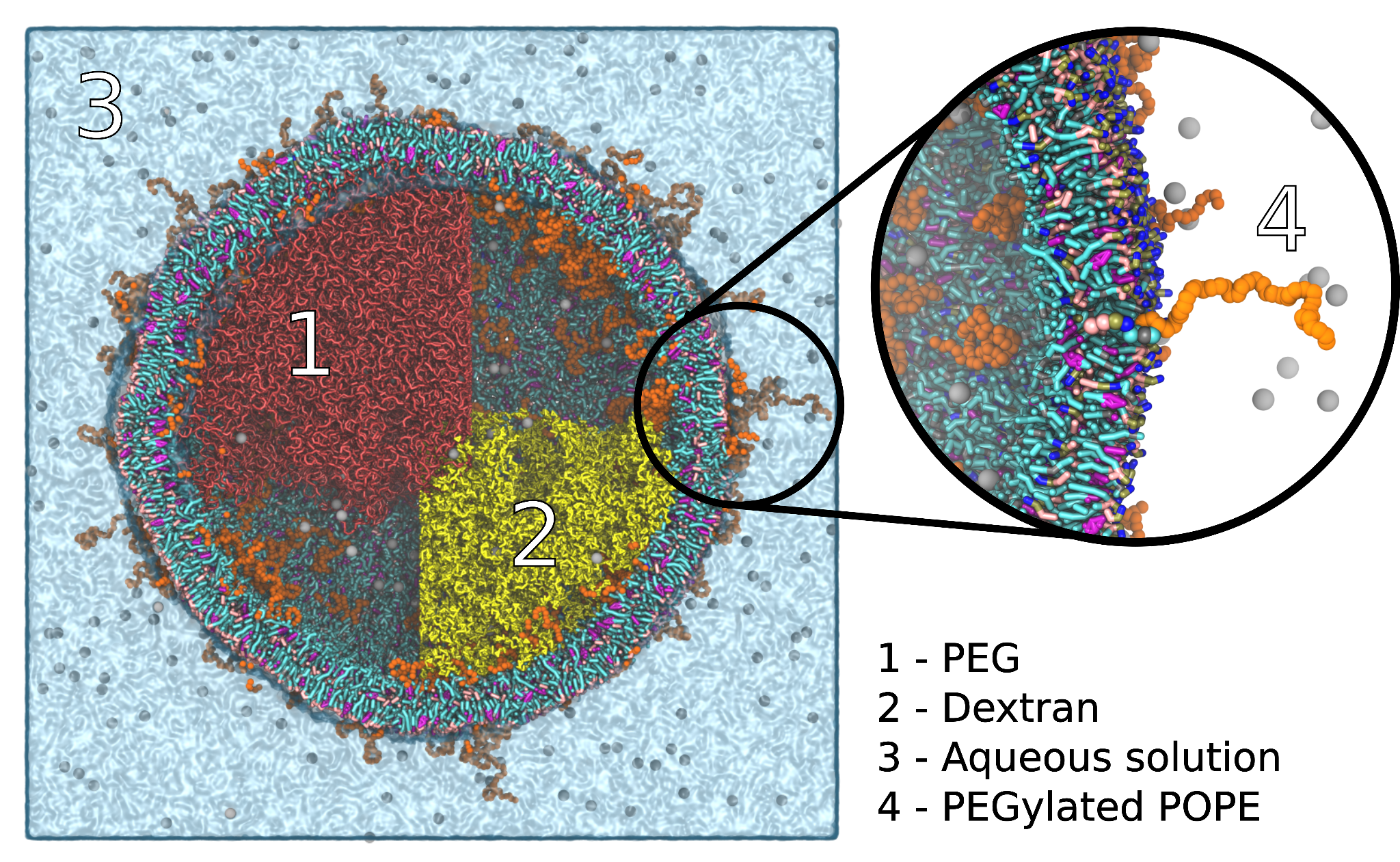
In this tutorial you will generate a model system for early life. In particular you will fill a vesicle with a phase separated mixture of PEO and Dextran (a carbohydrate polymer). This tutorial covers how to build phase separated systems, use pre-build coordinates from other programs, generate itp files for branched molecules, generate statistical distributions of polymers. The Tutorial comes in the form of a jupyter notebook (https://jupyter.org/). Make sure to have polyply installed before opening it. If you work within a virtual environment you will need to add the jupyter kernel before running the notebook. Instructions can be found in the notebook.
Download here.
[1] Grünewald, F., Alessandri, R., Kroon, P. C., Monticelli, L., Souza, P. C., & Marrink, S. J. (2021). Polyply: a python suite for facilitating simulations of (bio-) macromolecules and nanomaterials. arXiv preprint arXiv:2105.05890.


























































































































































