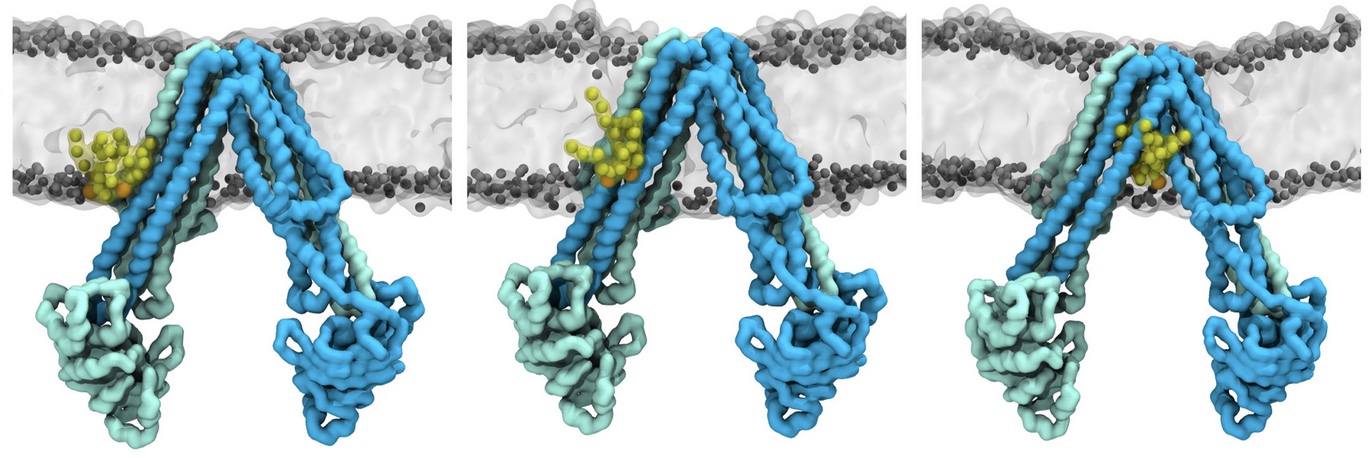
Lipid A binding to ABC transporter
- Details
- Last Updated: Monday, 17 October 2022 14:39
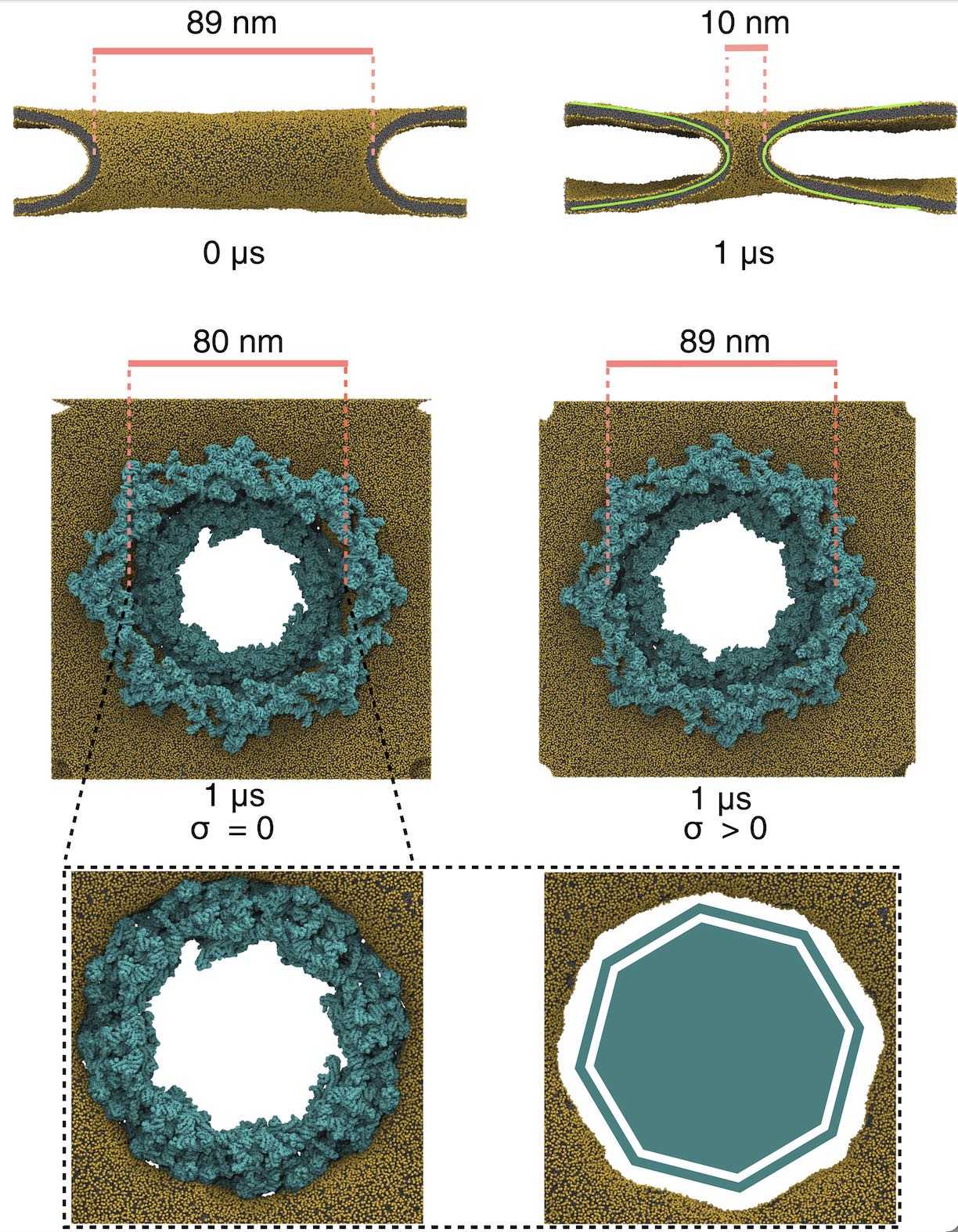
Molecular dynamics simulations of the ABC transporter MsbA, performed by Lars Schäfer and coworkers, revealed that extensive lateral portal opening is essential to provide access of its large natural substrate core lipid A to the binding cavity.
Cool Martini stuff ! For details, see Galazzo et al., Science Advances (2022) 8,41. DOI: 10.1126/sciadv.abn6845