Current rectification hemolysin
- Details
- Last Updated: Wednesday, 08 June 2022 12:41
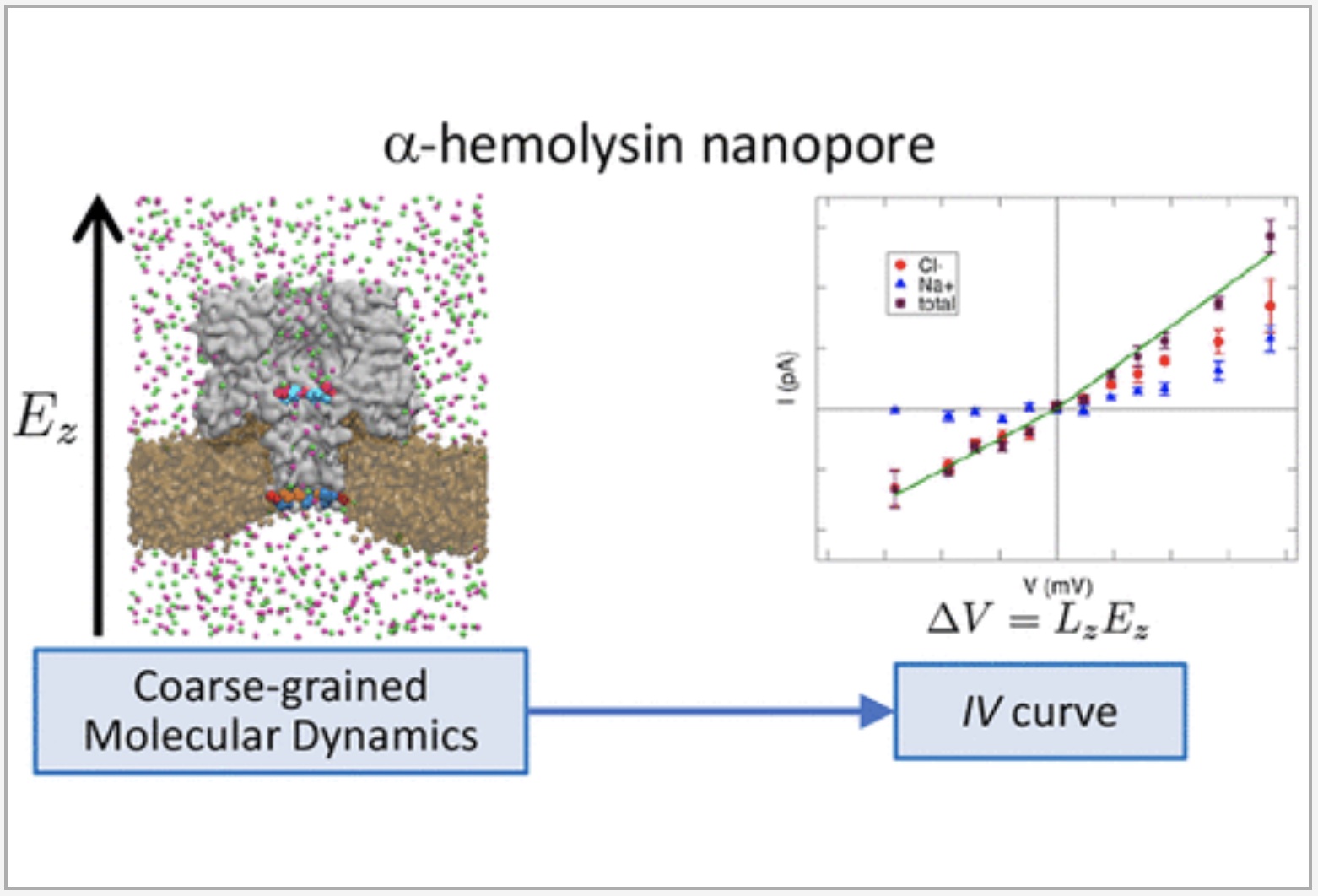 The group of Basdevant performed Martini simulations of the ionic transport through the α-hemolysin protein nanopore, using the polarizable water model. The electric potential difference applied experimentally was mimicked by the application of an electric field to the system. They were able to observe current asymmetry and anion selectivity, in agreement with previous studies and experiments, and identified the charged amino acids responsible for these current behaviors.
The group of Basdevant performed Martini simulations of the ionic transport through the α-hemolysin protein nanopore, using the polarizable water model. The electric potential difference applied experimentally was mimicked by the application of an electric field to the system. They were able to observe current asymmetry and anion selectivity, in agreement with previous studies and experiments, and identified the charged amino acids responsible for these current behaviors.
For details, see Dessaux et al., JPCB, 2022.



























































































































































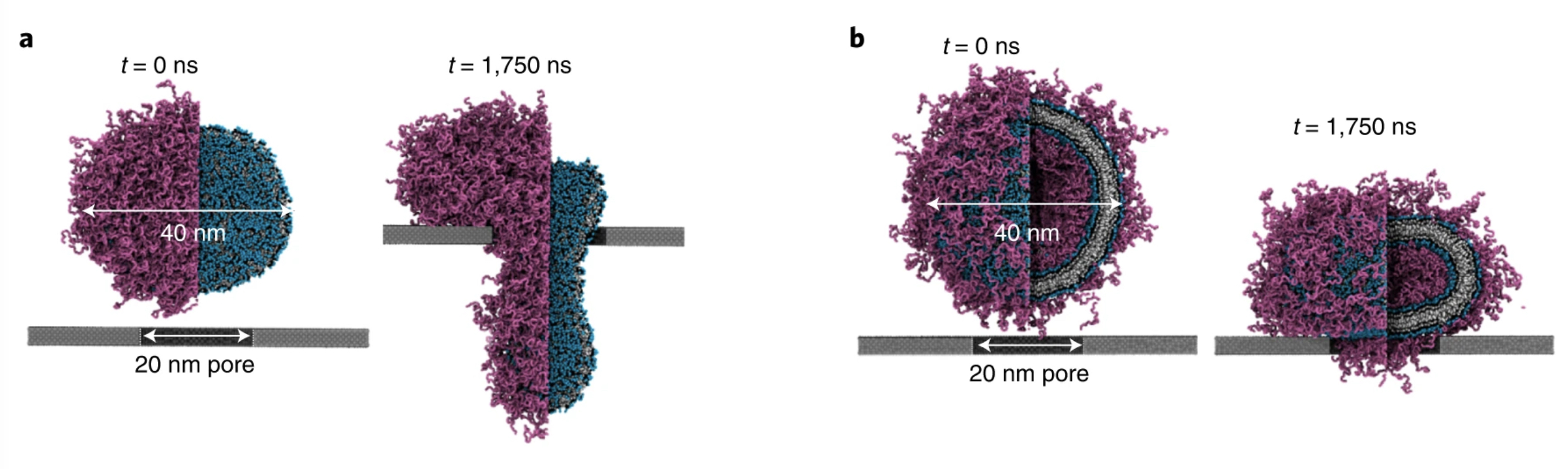
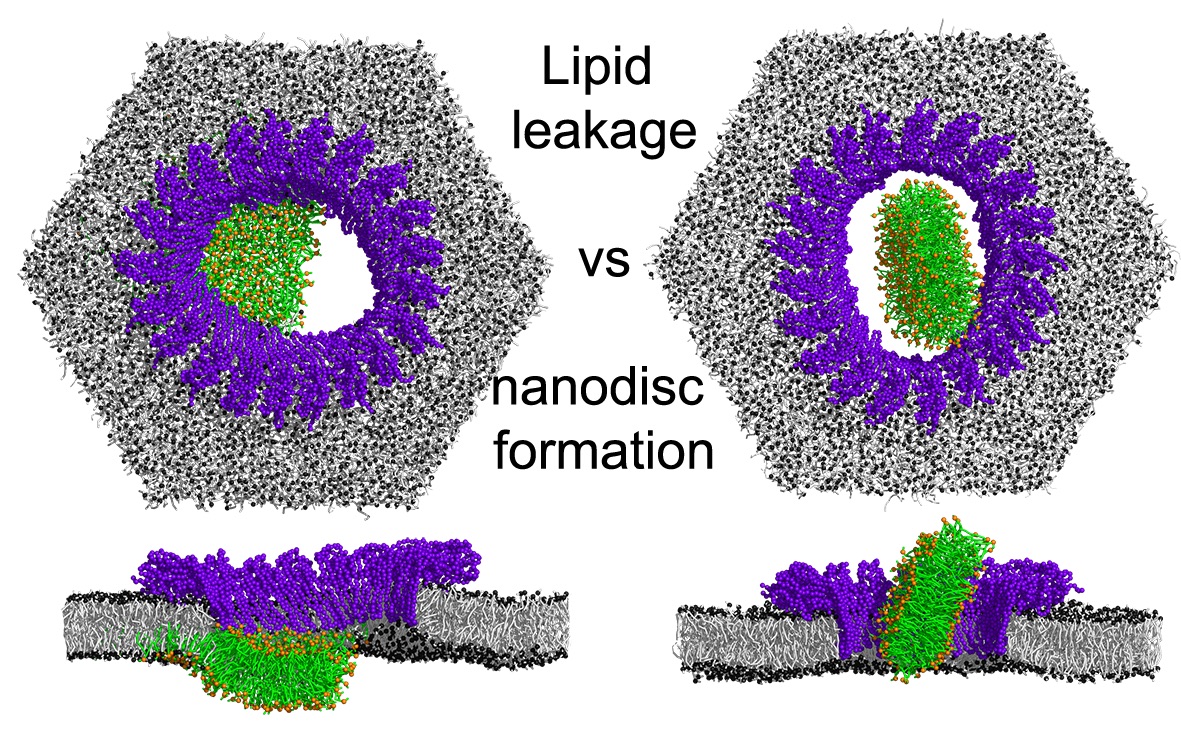 Simulations with Martini 3 reveal how lipids unplug pyroptotic pores formed by gasdermin-A3.
Simulations with Martini 3 reveal how lipids unplug pyroptotic pores formed by gasdermin-A3.
