Membrane permeability
- Details
- Last Updated: Monday, 28 March 2022 11:51
 General Purpose Coarse-Grained Force Field
General Purpose Coarse-Grained Force Field 

























































































































































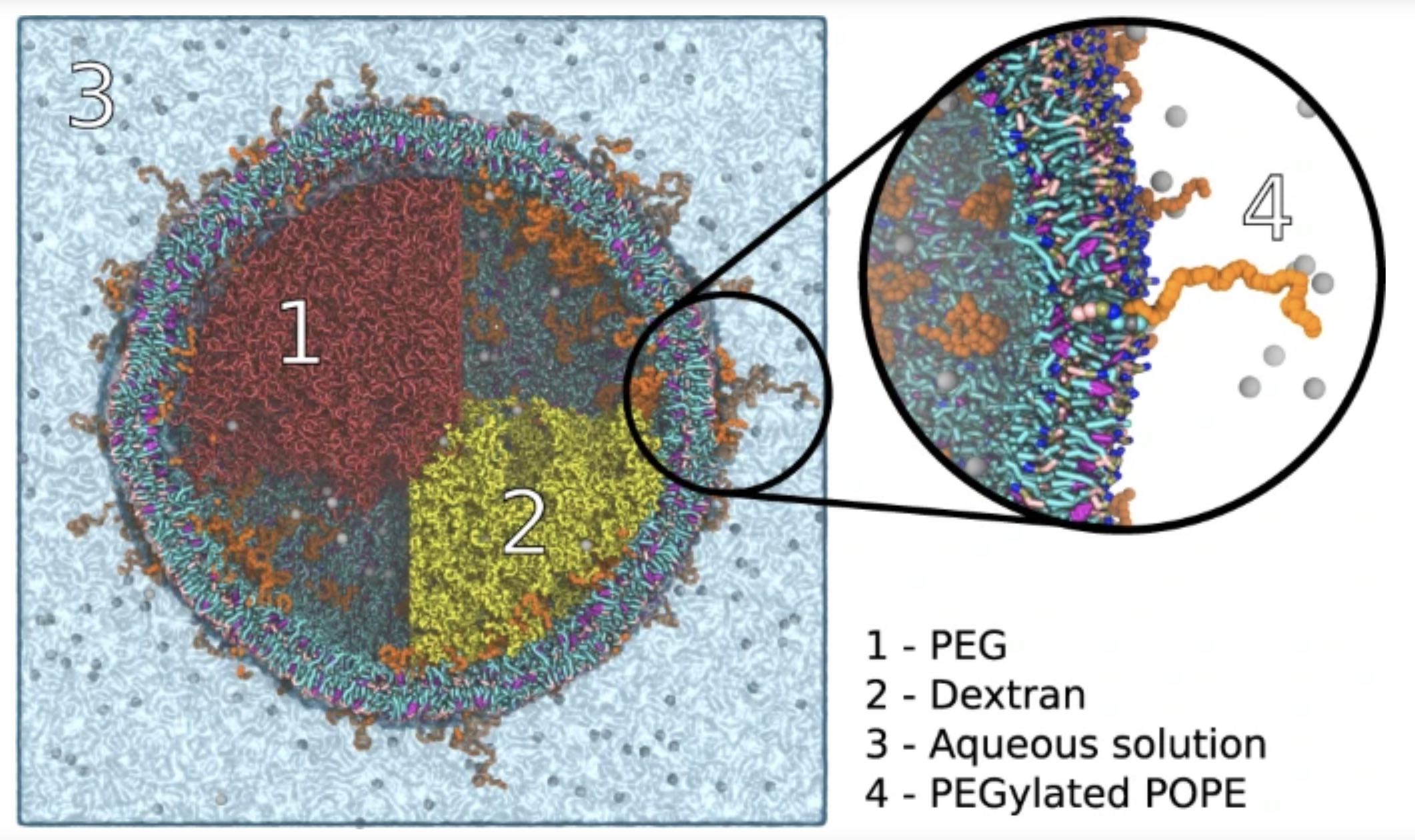 Generating input files and realistic starting coordinates for complex multi-component simulations is often a major bottleneck, especially for high throughput protocols. To eliminate this bottleneck, we present the polyply software suite that provides 1) a multi-scale graph matching algorithm designed to generate parameters quickly and for arbitrarily complex polymeric topologies, and 2) a generic multi-scale random walk protocol capable of setting up complex systems efficiently and independent of the target force-field or model resolution.
Generating input files and realistic starting coordinates for complex multi-component simulations is often a major bottleneck, especially for high throughput protocols. To eliminate this bottleneck, we present the polyply software suite that provides 1) a multi-scale graph matching algorithm designed to generate parameters quickly and for arbitrarily complex polymeric topologies, and 2) a generic multi-scale random walk protocol capable of setting up complex systems efficiently and independent of the target force-field or model resolution.
We demonstrate the power of polyply by setting up a number of complex systems, including a liquid-liquid phase separated polymer system inside a lipid vesicle. For details, see Grünewald et al., Nature Commun. 2022.
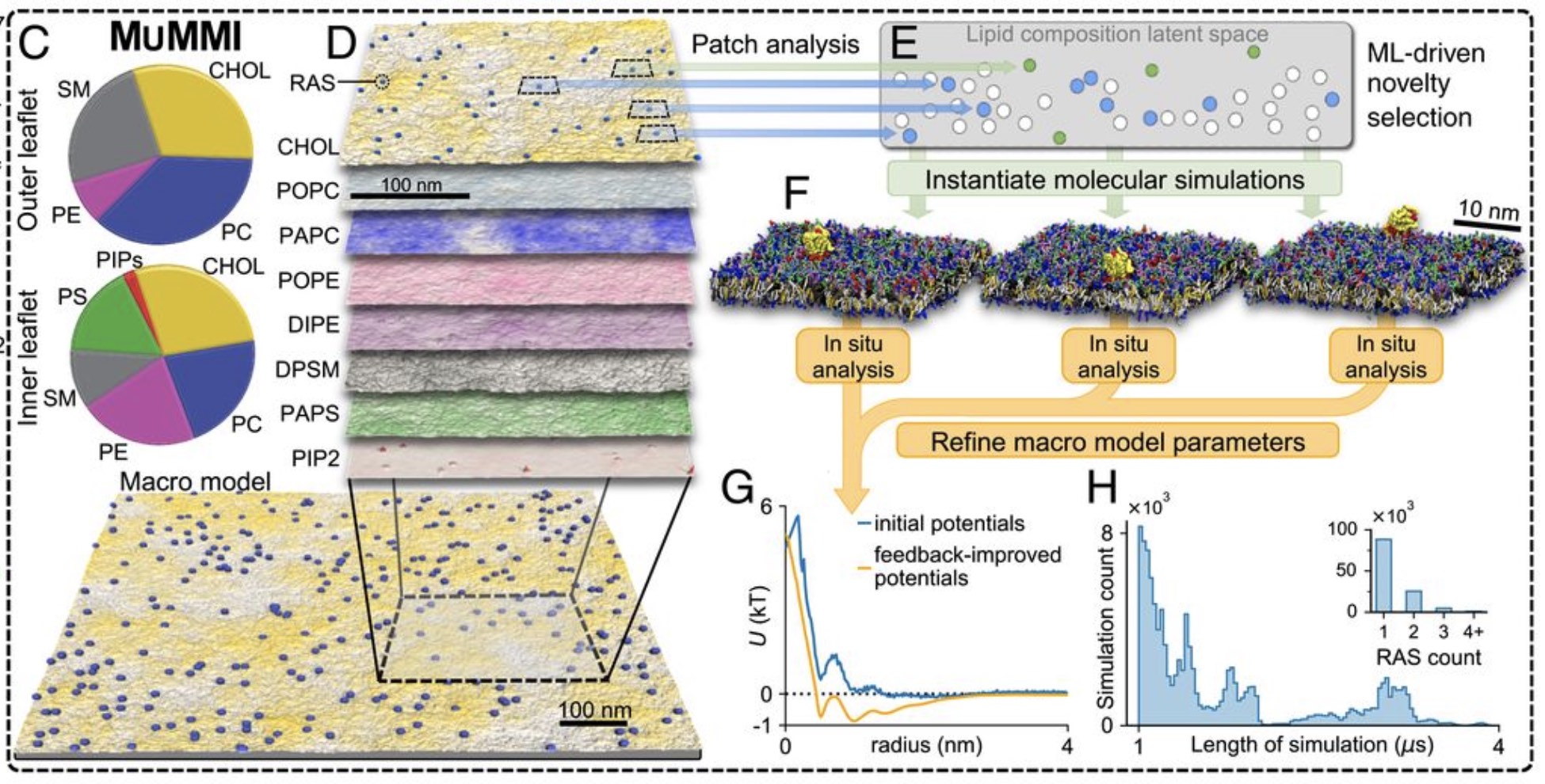
A huge consortium effort from the Livermore Lab on RAS dynamics, featuring a staggering 120,000 independent Martini simulations, has now been published: Ingolfsson et al., PNAS, 2022.
Impressive work based on the MuMMI (Multiscale Machine-learned Modeling Infrastructure) workflow.
 A good start of the new year: Our paper on how to parameterise small molecules with Martini 3 is now published !
A good start of the new year: Our paper on how to parameterise small molecules with Martini 3 is now published !
You can read about best practices on chosing bead types, handling constraints, and selecting virtual sites for planar compounds, as well as find a large table with best bead types for a wide selection of chemical fragments and numerous validated topologies for small compounds.
Alessandri et al., Adv. Theory & Simul. 2100391. https://doi.org/10.1002/adts.202100391
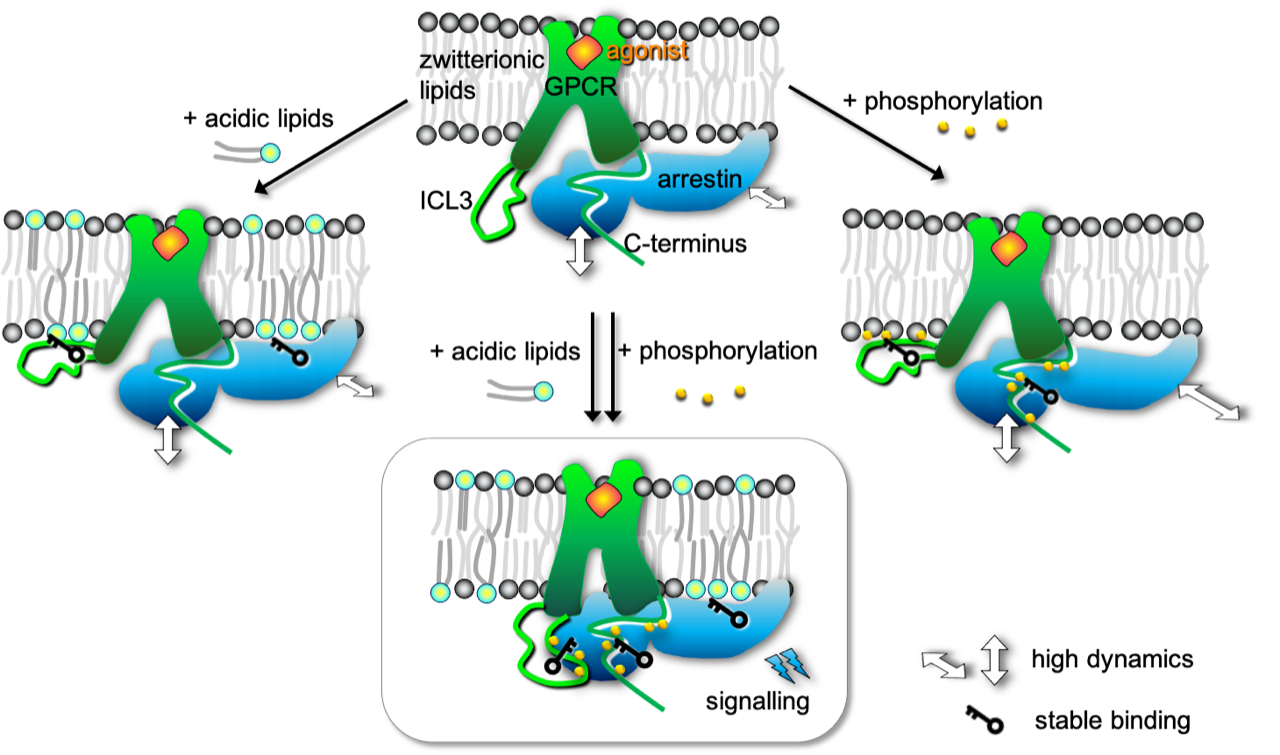 Martini-based simulations capture the spontaneous binding of beta-arrestin2 to beta2-adrenergic receptor in a similar pose as the crystallographically resolved structure of rhodopsin/arresin-1. Cool !
Martini-based simulations capture the spontaneous binding of beta-arrestin2 to beta2-adrenergic receptor in a similar pose as the crystallographically resolved structure of rhodopsin/arresin-1. Cool !
See the paper from Pluhackova et al.: https://doi.org/10.3389/fcell.2021.807913

From the lab of Tajkhorshid: a protocol for setting up simulations of cell-scale membrane envelopes, using Martini.
Paving the way for whole cell simulations !
See Vermaas et al., JCIM, online, for details.