Membranes in situ
- Details
- Last Updated: Wednesday, 15 May 2024 15:02

Why not include the rest of the cell in your next membrane simulation ?
GoMartini virtual sites
- Details
- Last Updated: Wednesday, 17 April 2024 09:52
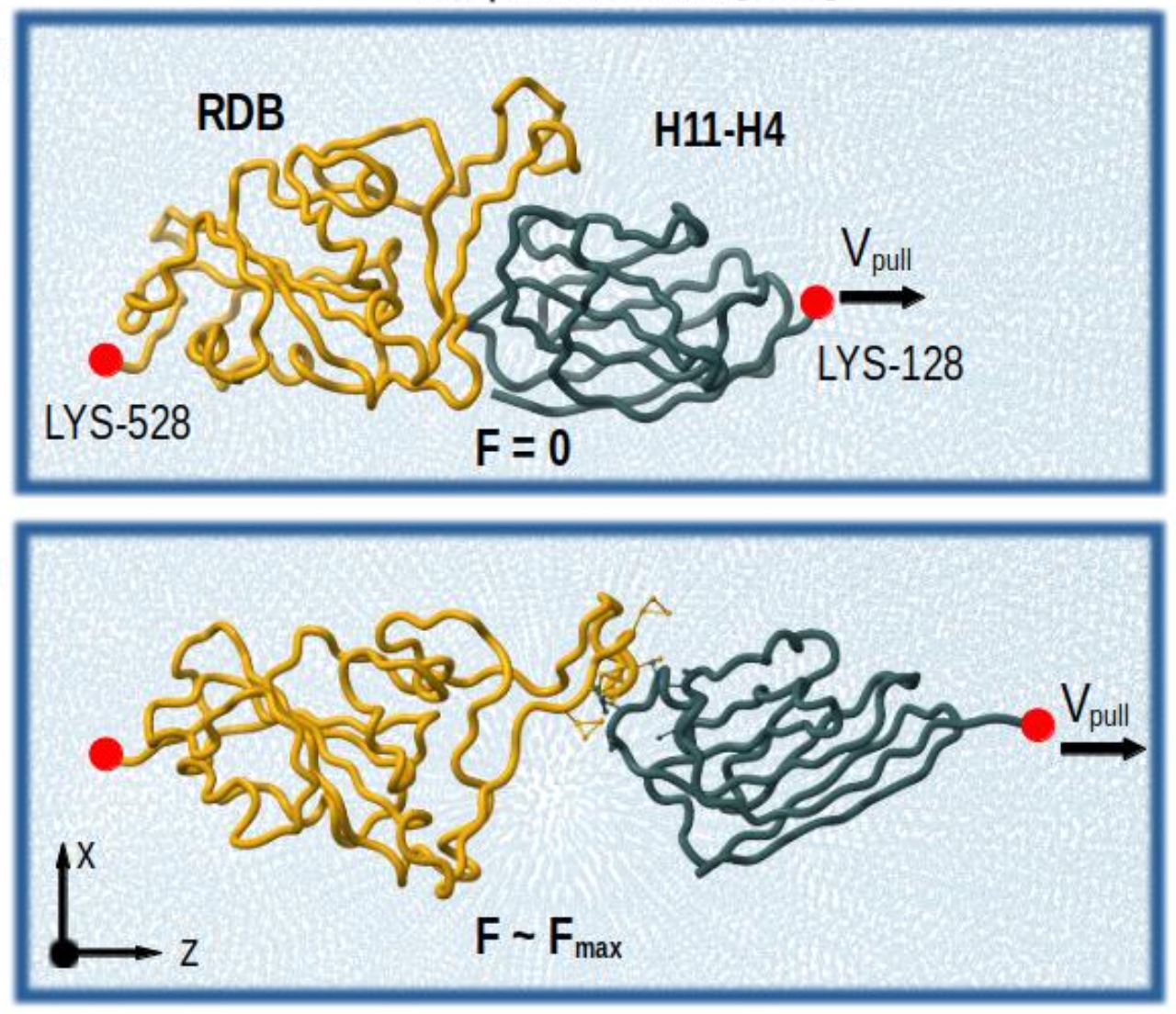
A brand new paper introducing an enhanced GōMartini model, combining a virtual-site implementation of Gō models with Martini 3:
https://www.biorxiv.org/content/10.1101/2024.04.15.589479v1
This work demonstrates the capabilities of the model in diverse case studies, ranging from protein-membrane binding to protein-ligand interactions and AFM force profile calculations. The model is also versatile, as it can address recent inaccuracies reported in the Martini protein model.
Lastly, the paper discusses the advantages, limitations, and future perspectives of the Martini 3 protein model and its combination with Gō models.
Building complex membranes
- Details
- Last Updated: Tuesday, 16 April 2024 07:25
SARS-CoV-2 model
- Details
- Last Updated: Friday, 08 December 2023 13:33
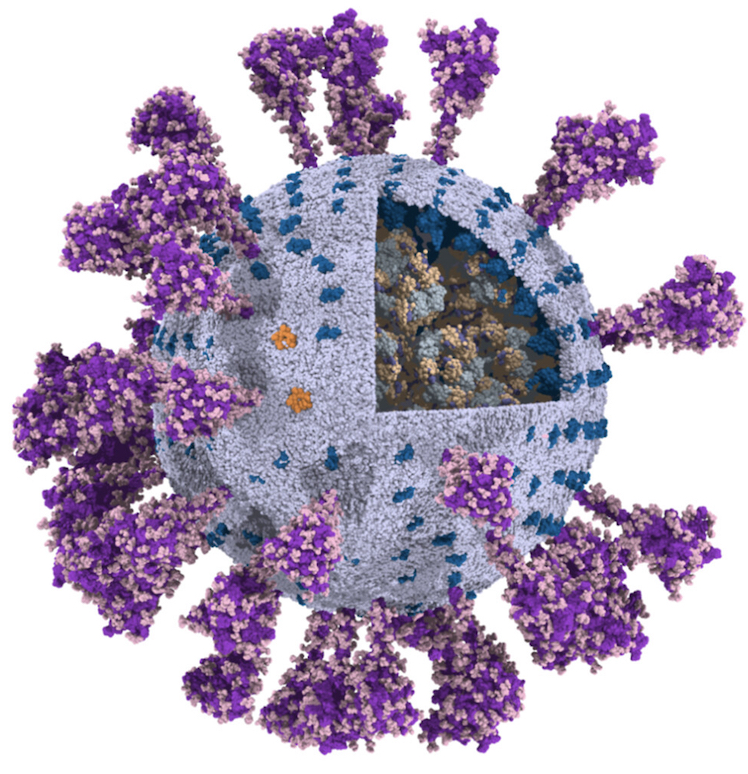
The group of Chen Song has published a fully-atomistic model of the SARS-CoV-2 envelope, including N-bound RNA segments, obtained after backmapping from a 500 ns Martini simulation (final configuration shown on the left).
For details, see: Wang et al., Quant. Biology, 2023.
Martini condensates
- Details
- Last Updated: Saturday, 25 November 2023 20:20

Multiscale simulations reveal TDP-43 molecular-level interactions driving condensation.
For details, check the paper from Ingolfsson et al. that just appeared in Biophysical Journal:


























































































































































